Publications
2024
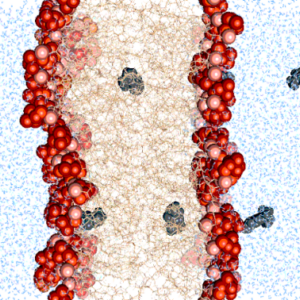
We find that commonly used photoresist molecules can pass the cell membrane by passive diffusion. This may be important to consider for bioprinting applications.
Diedrich, L.#, Brosz, M., Abele, T., Urbanek, N., Steinke, S., Gräter, F.*, Göpfrich, K.*, Aponte-Santamaría, C.* (2024) Energetics and Kinetics of Membrane Permeation of Photoresists for Bioprinting. Advanced Functional Materials. DOI: 10.1002/adfm.202315731
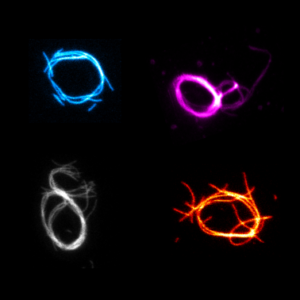
We build contractile rings with DNA nanotechnology as a first step towards a synthetic division machninery for synthetic cells:
Illig, M.#, Jahnke, K.#, Weise, L.P.#, Scheffold, M., Mersdorf, U., Drechsler, H., Zhang, Y., Diez, S.*, Kierfeld, J.* & Göpfrich, K.* (2024) Triggered contraction of self-assembled micron-scale DNA nanotube rings. Nature Communications 15, 2307. DOI: 10.1038/s41467-024-46339-z
2023

A reflection on our efforts to engineer a cell (in German):
Göpfrich, K. (2023) Molecularer Balanceakt: Synthetische Biologie schafft künstliches Leben, Ruperto Carola 22. DOI: 10.17885/heiup.ruca.2023.22.24822
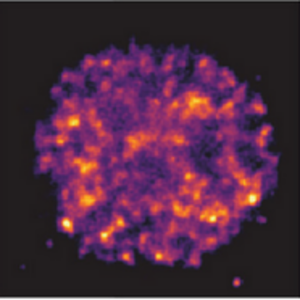
We collaborated with the Hilbert and Zaburdaev groups to mimic kinetics of transcriptional clusters in the cell nucleus with DNA droplets:
Tschurikow, X., Gadzekpo, A., Tran, M.P., Chatterjee, R., Sobucki, M., Zaburdaev, V., Göpfrich, K. & Hilbert, L. (2023) Amphiphiles Formed from Synthetic DNA-Nanomotifs Mimic the Stepwise Dispersal of Transcriptional Clusters in the Cell Nucleus, Nano Letters. DOI: 10.1021/acs.nanolett.3c01301
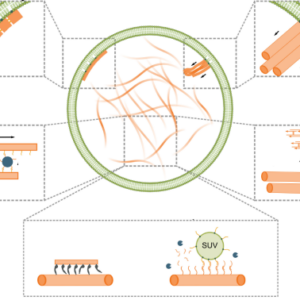
We discuss advances in engineering mimics of cytoskeletons with DNA nanotechnology:
Jahnke, K. & Göpfrich, K. (2023). Engineering DNA-based cytoskeletons for synthetic cells. Interface Focus 13, 20230028. DOI: 10.1098/rsfs.2023.0028
We take a closer look at the new world of RNA origami. This is why we are excited about it:
Poppleton, E., Urbanek, N., Chakraborty, T., Griffo, A., Monari, L. & Göpfrich, K. (2023). RNA origami: design, simulation and application. RNA biology 20, 510-524. DOI: 10.1080/15476286.2023.2237719
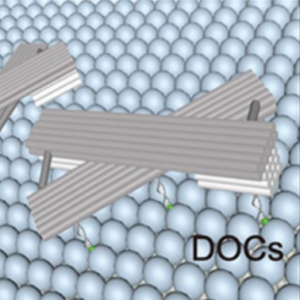
We switch the conformation of DNA origami lattices on lipid vesicles and thereby control vesicle morphology.
Yang, J., Jahnke, K., Xin, L., Jing, X., Zhan, P., Peil, A., Griffo, A., Škugor, M., Yang, D., Fan, S., Göpfrich, K.*, Yan, H.*, Wang, P.* & Liu, N. (2023). Modulating Lipid Membrane Morphology by Dynamic DNA Origami Networks. Nano Letters. DOI: acs.nanolett.3c00750
I report about a recent study which shows how mechanochemical feedback loops drive the persistent motion of liposomes (News & Views).
Göpfrich, K. (2023) Synthetic cells on the move. Nature Physics. DOI: 10.1038/s41567-023-02137-w
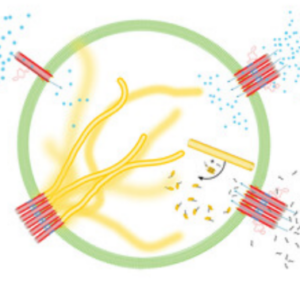
We engineer signal transduction in synthetic cells: An external chemical signal triggeres the mechanical remodelling of a DNA cytoskeleton in a GUV.
Jahnke, K., Illig, M., Scheffold, M., Tran, M.P., Mersdorf, U. & Göpfrich, K. (2023) DNA Origami Signaling Units Transduce Chemical and Mechanical Signals in Synthetic Cells. Advanced Functional Materials 2301176. DOI: 10.1002/adfm.202301176
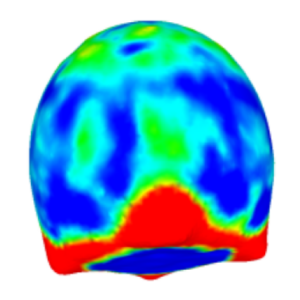
We provide a software package to extract quantitative data from z-stacks of vesicles, droplets and cells.
Dreher, Y., Niessner, J., Fink, A. & Göpfrich, K. (2023). GeoV: An Open-Source Software Package for Quantitative Image Analysis of 3D Vesicle Morphologies. Advanced Intelligent Systems, 2300170. DOI: 10.1002/aisy.202300170
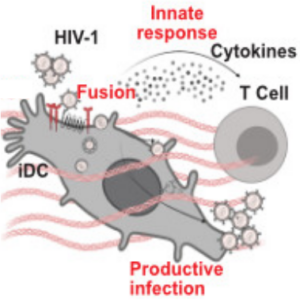
In collaboration with the group of Oliver Fackler, we take a closer look at how the cell culture environment of immature dendritic cells influences HIV-1 infection. We contributed microfluidic experiments (real-time deformability cytometry) to quantify cell stiffness.
Gallucci, L., Abele, T., Fronza, R., Stolp, B., Laketa, V., Sid Ahmed, S., Flemming, A., Müller, B., Göpfrich, K. & Fackler, O. T. (2023). Tissue‐like environments shape functional interactions of HIV‐1 with immature dendritic cells. EMBO reports, e56818. DOI: 10.15252/embr.202356818
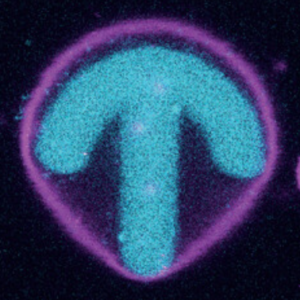
In collaboration with the group of Ada Cavalcanti-Adam, we adhere GUVs to photopatterend surfaces and study the organization of internal actin:
A. Fink, C. R. Doll, A. Y. Relimpio, Y. Dreher, J. P. Spatz, K. Göpfrich, and E. A. Cavalcanti-Adam* (2023). Extracellular Cues Govern Shape and Cytoskeletal Organization in Giant Unilamellar Lipid Vesicles. ACS Synthetic Biology. DOI: 10.1021/acssynbio.2c00516
2022
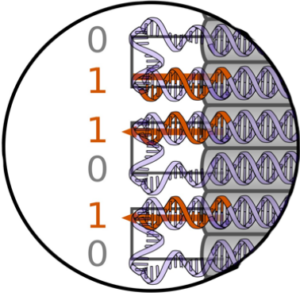
We explore properties of genotype-phenotype maps with DNA origami polyominos:
Dreher, Y., Fichtler, J., Karfusehr, C., Jahnke, K., Xin, Y., Keller, A., & Göpfrich, K. (2022). Genotype-phenotype mapping with polyominos made from DNA origami tiles. Biophysical Journal. DOI: 10.1016/j.bpj.2022.09.006
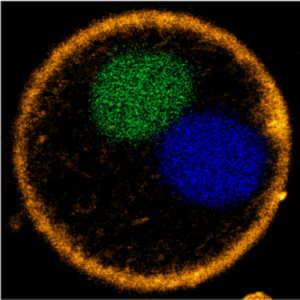
We demonstrate a DNA segregation module for synthetic cells by segregating DNA droplets inside GUVs:
M. P. Tran, R. Chatterjee, Y. Dreher, J. Fichtler, K. Jahnke, L. Hilbert, V. Zaburdaev*, K. Göpfrich*. A DNA segregation module for synthetic cells (2022). Small 2202711. DOI: 10.1002/smll.202202711
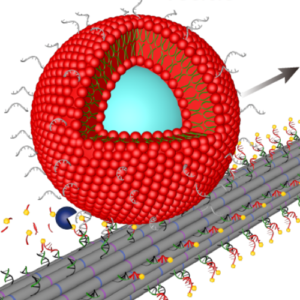
We build DNA-based mimicks of cytoskeletal filaments capable of reversible assembly and cargo transport:
P. Zhang, K. Jahnke*, N. Liu*, K. Göpfrich*, Functional DNA-based cytoskeletons for synthetic cells (2022). Nature Chemistry. DOI: 10.1038/s41557-022-00945-w
Highlighted in Nature; Press release (German & English): Another step towards synthetic cells
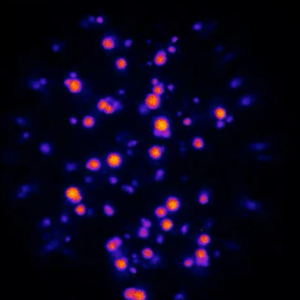
In collaboration with the group of Job Boekhoven at TU Munich we watch active droplets inside microfluidic droplets:
A. M. Bergmann, C. Donau, F. Späth, K. Jahnke, K. Göpfrich*, J. Boekhoven*, Evolution and Single-Droplet Analysis of Fuel-Driven Compartments by Droplet-Based Microfluidics (2022). Angewandte Chemie. DOI: 10.1002/anie.202203928
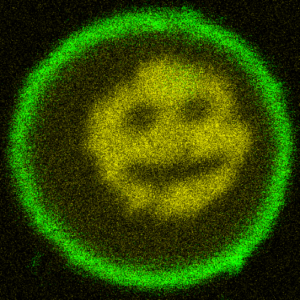
We turn DNA into a photoresist and print micron-sized structures inside lipid vesicles:
T. Walther, K. Jahnke, T. Abele, K. Göpfrich*, Printing and Erasing of DNA‐Based Photoresists Inside Synthetic Cells (2022). Advanced Functional Materials, 2200762. DOI: 10.1002/adfm.202200762
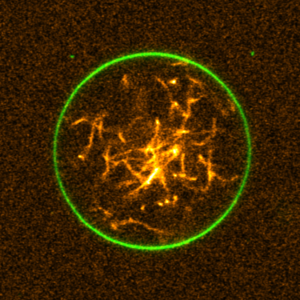
We engineered a DNA-cytoskeleton, which can be assembled and disassembled by light, deform lipid vesicles from inside and can be bundled into ring-like structures:
K. Jahnke, V. Huth, U. Mersdorf, N. Liu, K. Göpfrich*, Bottom-Up Assembly of Synthetic Cells with a DNA Cytoskeleton. ACS Nano (2022). DOI: 10.1021/acsnano.1c10703
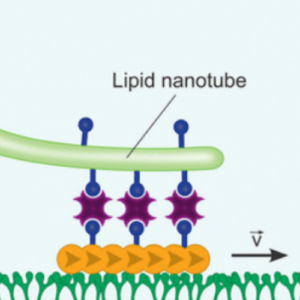
We used molecular motors to pull out lipid nanotubes from cellular membranes:
K. Jahnke, S. J. Maurer, C. Weber, J. E. Hernandez Bücher, A. Schoenit, E. D’Este, E. A. Cavalcanti-Adam, K. Göpfrich*, Actomyosin-Assisted Pulling of Lipid Nanotubes from Lipid Vesicles and Cells. Nano Letters 22, 3, 1145–1150 (2022). DOI: 10.1021/acs.nanolett.1c04254
2021
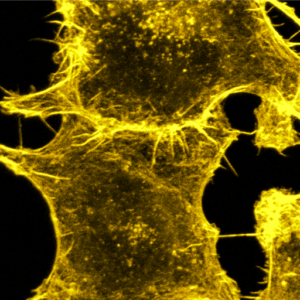
We tune cell-cell adhesion strength in epithelial cell collectives with DNA nanotechnology:
A. Schoenit, C. Lo Giudice, N. Hahnen, D. Ollech, K. Jahnke, K. Göpfrich*, E. A. Cavalcanti-Adam*, Tuning Epithelial Cell–Cell Adhesion and Collective Dynamics with Functional DNA-E-Cadherin Hybrid Linkers. Nano Letters (2021). DOI: 10.1021/acs.nanolett.1c03780
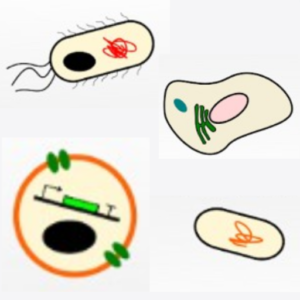
Building a synthetic cell is a community efford. This is how we envision this community to grow and thrive:
O. Staufer, J. De Lora, E. Bailoni, A. Bazrafshan, A. S. Benk, K. Jahnke, Z. A. Manzer, L. Otrin, T. D. Pérez, J. Sharon, J. Steinkühler, K. P. Adamala, B. Jacobson, M. Dogterom, K. Göpfrich, D. Stefanovic, S. R. Atlas, M. Grunze, M. R. Lakin, A. P. Shreve, J. P., Spatz, G. P., López, Building a community to engineer synthetic cells and organelles from the bottom-up. Elife 10, e7556 (2021). DOI: 10.7554/eLife.73556
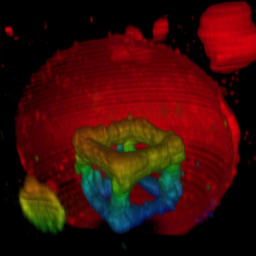
How can you build up structures on the inside of synthetic cells? With two-photon 3D laser printing:
T. Abele, T. Messer, K. Jahnke, M. Hippler, M. Bastmeyer, M. Wegener & K. Göpfrich. Two-photon 3D laser printing inside synthetic cells, Advanced Materials (2021). DOI: 10.1002/adma.202106709
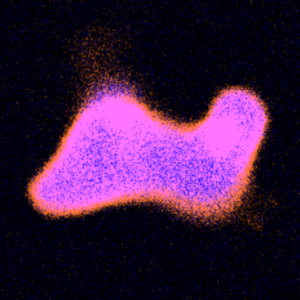
We divide DNA-containing giant unilamellar lipid vesicles with light-triggered lipid peroxidation:
Y. Dreher, K. Jahnke, M. Schröter & K. Göpfrich. Light-Triggered Cargo Loading and Division of DNA-Containing Giant Unilamellar Lipid Vesicles, Nano Letters (2021). DOI: 10.1021/acs.nanolett.1c00822
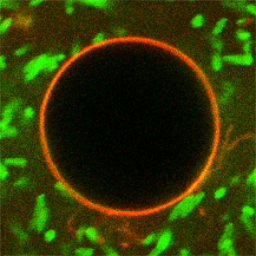
We use top-down engineered E. coli as actuators in bottom-up assembled synthetic cells:
K. Jahnke, N. Ritzmann, J. Fichtler, A. Nitschke, Y. Dreher, T. Abele, G. Hofhaus, I. Platzman, R. Schröder, D. Müller, J. Spatz & K. Göpfrich. Proton gradients from light-harvesting E. coli control DNA assemblies for synthetic cells, Nature Communications 12, 3967, 2021. https://doi.org/10.1038/s41467-021-24103-x
Press release: https://www.mr.mpg.de/14524114/synthetic-biology-bottom-up-meets-top-down
Behind the Paper: https://bioengineeringcommunity.nature.com/posts/synthetic-biology-bottom-up-meets-top-down
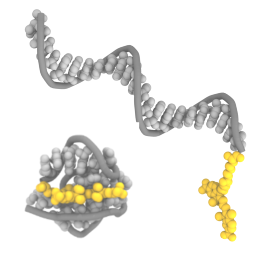
We show with experiments and molecular dynamics simulations that the choice of fluorophores matters for dynamic DNA nanostructures:
K. Jahnke, H. Grubmüller, M. Igaev & K. Göpfrich. Choice of fluorophore affects dynamic DNA nanostructures, Nucleic Acids Research gkab201, 2021 https://doi.org/10.1093/nar/gkab201
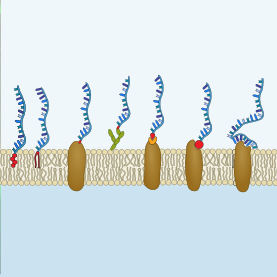
We provide tips, tricks and tools for the functionalization of cellular membranes with DNA nanotechnology and review recent applications:
A. Schönit, A. Cavalcanti-Adam & K. Göpfrich. Functionalization of Cellular Membranes with DNA Nanotechnology, Trends in Biotechnology, 2021. https://doi.org/10.1016/j.tibtech.2021.02.002
2020
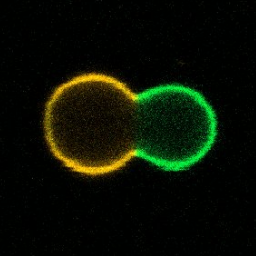
We divide phase-separated GUVs with enzymatic reactions or locally with light and describe the process with an analytical model:
Y. Dreher, K. Jahnke, E. Bobkova, J.P. Spatz & K. Göpfrich. Division and regrowth of phase‐separated giant unilamellar vesicles, Angewandte Chemie, 2020. https://doi.org/10.1002/ange.202014174

We share some methods and protocols for the generation of DNA-based membrane pores in a bookchapter:
K. Göpfrich, A. Ohmann & U. F. Keyser. Design and Assembly of Membrane-Spanning DNA Nanopores. In: Nanopore Technology (Ed. M. Fahie), 2020. ISBN 978-1-0716-0806-7
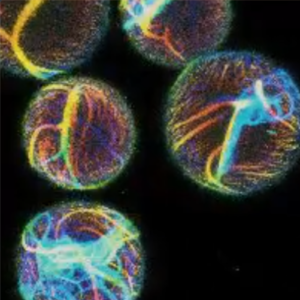
We found that actin-containing droplet show directional motion due to the Marangoni effect:
B. Haller, K. Jahnke, M. Weiss, K. Göpfrich*, I. Platzman* & J. P. Spatz*. Autonomous Directional Motion of Actin-Containing Cell-Sized Droplets, Advanced Intelligent Systems, 2020. https://doi.org/10.1002/aisy.202000190
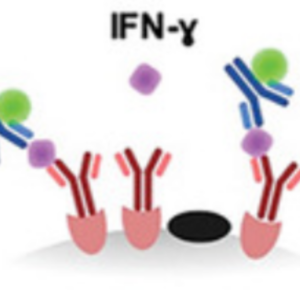
We contributed to an assay to evaluate cell cytotoxicity and cytokine release on a single-cell level:
S. Antona, T. Abele, K. Jahnke, Y. Dreher, K. Göpfrich, I. Platzman & J. P. Spatz. Droplet‐Based Combinatorial Assay for Cell Cytotoxicity and Cytokine Release Evaluation, Advanced Functional Materials, 2003479, 2020. https://doi.org/10.1002/adfm.202003479

Cholesterol-tagged DNA does not only self-assemble into lipid membranes but also polymersomes – under the right conditions:
R. Luo, K. Göpfrich, I. Platzman & J. P. Spatz. DNA-Based Assembly of Multi-Compartment Polymersome Networks, Advanced Functional Materials, 2003480, 2020. https://doi.org/10.1002/adfm.202003480

We interface DNA nanotechnology and actin networks and show that light-triggered contaction breaks the symmetry:
K. Jahnke, M. Weiss, C. Weber, I. Platzman*, K. Göpfrich* & J. P. Spatz*. Engineering Light‐Responsive Contractile Actomyosin Networks with DNA Nanotechnology, Advanced Biosystems 2020. https://doi.org/10.1002/adbi.202000102

We used DNA functionalization and electrocoalescence to filter the content of microfluidic droplets:
C. Frey, K. Göpfrich, S. Pashapour, I. Platzman & J. P. Spatz. Electrocoalescence of Water-in-Oil Droplets with a Continuous Aqueous Phase: Implementation of Controlled Content Release. ACS Omega 2020. https://doi.org/10.1021/acsomega.0c00344

We reconfigure plasmonic DNA origami in microfluidic droplets:
K. Göpfrich*, M. J. Urban, C. Frey, I. Platzman, J. P. Spatz* & N. Liu*. Dynamic Actuation of DNA-Assembled Plasmonic Nanostructures in Microfluidic Cell-Sized Compartments. Nano Letters 20, 1571-1577, 2020. https://doi.org/10.1007/978-981-13-9791-2_11
2019

We contributed a bookchapter on DNA nanopores:
K. Göpfrich & U. F. Keyser. DNA Nanotechnology for Building Sensors, Nanopores and Ion-Channels. In: Biological and Bio-inspired Nanomaterials, 2019. https://doi.org/10.1007/978-981-13-9791-2_11

Our view on synthetic biology published in Heidelberg University’s research magazine Ruperta Carola (mostly in German):
K. Göpfrich, I. Platzman & J. P. Spatz. Aus dem Baukasten der molekularen Ingenieure. Auf dem Weg zur synthetischen Zelle, Ruperto Carola, 106-113 2019. [PDF]

We show that single-stranded DNA overhangs can wrap around a cholesterol-tag and thereby prevent aggregation of cholesterol-modified DNA nanostructures:
A. Ohmann, K. Göpfrich, H. Joshi, R. F. Thompson, D. Sobota, N. A. Ranson, A. Aksimentiev & U. F. Keyser. Controlling aggregation of cholesterol-modified DNA nanostructures. Nucleic Acids Research 47, 11441–11451, 2019. https://doi.org/10.1093/nar/gkz914
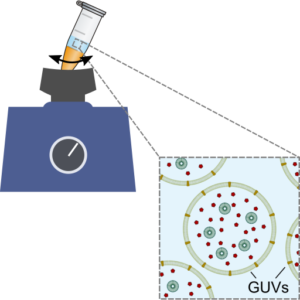
We establish a method for the formation of giant unilamellar vesicles for the assembly of synthetic cells, offering straight-forward encapsulation of content:
K. Göpfrich, B. Haller, O. Staufer, Y. Dreher, U. Mersdorf, I. Platzman & J. P. Spatz, One-Pot Assembly of Complex Giant Unilamellar Vesicle-Based Synthetic Cells. ACS Synthetic Biology, 2019. https://doi.org/10.1021/acssynbio.9b00034
Video protocol illustrating the method: https://youtu.be/vOPp97toPAw
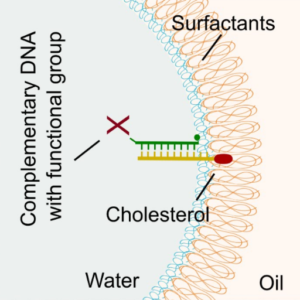
We demonstrate a universal strategy for the functionalization of microfluidic droplets by attaching reactive groups and components to cholesterol-tagged DNA handles:
K. Jahnke, M. Weiss, C. Frey, S. Antona, J.-W. Janiesch, I. Platzman, K. Göpfrich* & J. P. Spatz*, Programmable Functionalization of Surfactant-Stabilized Microfluidic Droplets via DNA-Tags. Advanced Functional Materials, 2019. https://doi.org/10.1002/adfm.201808647
2018

By tailoring the charge density at the interface of microfluidic droplets, we control the transition between multicompartment systems and GUVs:
B. Haller, K. Göpfrich, M. Schröter, J.-W. Janiesch, I. Platzman & J. P. Spatz, Charge-controlled microfluidic formation of lipid-based single- and multicompartment systems. Lab on a Chip, 2018. https://doi.org/10.1039/C8LC00582F

In this review, we discuss how microfluidics and DNA nanotechnology can be used as tools to assemble complex synthetic cells.
K. Göpfrich*, I. Platzman* & J. P. Spatz*, Mastering Complexity: Towards Bottom-up Construction of Multifunctional Eukaryotic Synthetic Cells. Trends in Biotechnology, 2018. https://doi.org/10.1016/j.tibtech.2018.03.008
Watch a short video about our review here.
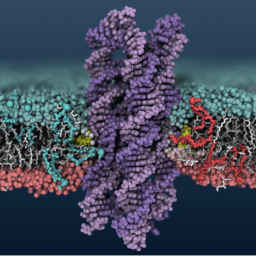
We demonstrate that membrane-spanning DNA nanopores are not just mimics of ion channels: They can also transport flip lipids from one bilayerleaflet to the other, like natural scramblases.
A. Ohmann, C.-Y. Li, C. Maffeo, K. Al Nahas, K. N. Baumann, K. Göpfrich, J. Yoo, U. F. Keyser, A. Aksimentiev, Outperforming nature: synthetic enzyme built from DNA flips lipids of biological membranes at record rates. Nature Communications, 2018. https://doi.org/10.1038/s41467-018-04821-5
In the news in c&en.
2017

Kerstin’s PhD thesis in the group of Prof. Ulrich F. Keyser at the University of Cambridge, on the assembly synthetic membrane pores from DNA.
K. Göpfrich, Rational Design of DNA-Based Lipid Membrane Pores. PhD Thesis, 2017. https://doi.org/10.17863/CAM.15517
Thanks to Gates Cambridge, the Winton Programme for the Physics of Sustainability and the Oppenheimer Trust for their generous support.
2016
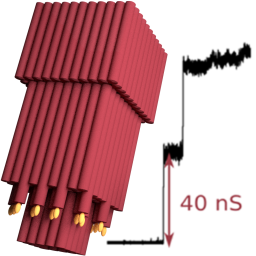
We built the largest man-made pore in lipid membranes to date and determine its conductance properties with single-molecule experiments and molecular dynamics simulations:
K. Göpfrich, C.-Y. Li, M. Ricci, S. P. Bhamidimarri, J. Yoo, B. Gyenes, A. Ohmann, M. Winterhalter, A. Aksimentiev & U. F. Keyser, Large-Conductance Transmembrane Porin Made from DNA Origami, ACS Nano, 2016. http://pubs.acs.org/doi/abs/10.1021/acsnano.6b03759
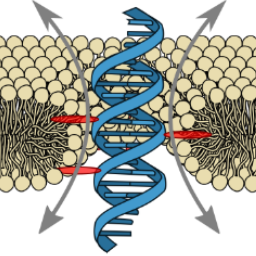
Ion conduction pathways across membranes can be lined by the lipids themselves. We demonstrated the formation of stable DNA-lipid pores induced by a single transmembrane-spanning DNA duplex:
K. Göpfrich, C.-Y. Li, C.-Y., I. Mames, S. P. Bhamidimarri, M. Ricci, J. Yoo, A. Mames, A. Ohmann, M. Winterhalter, E. Stulz, A. Aksimentiev & U. F. Keyser, Ion channels made from a single membrane-spanning DNA duplex. Nano Letters, 2016. http://pubs.acs.org/doi/abs/10.1021/acs.nanolett.6b02039
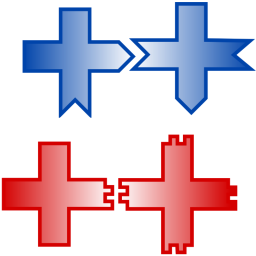
We study transitions from bound to unbound cluster growth using computational models and DNA-tile self-assembly experiments:
S. Tesoro, K. Göpfrich, T. Kartanas, U. F. Keyser & S. E. Ahnert. Non-deterministic self-assembly with asymmetric interactions can lead to tunable self-limiting cluster growth. Physical Review E, 2016. http://journals.aps.org/pre/abstract/10.1103/PhysRevE.94.022404
2015

We created the smallest membrane-inserting DNA nanostructure to date, approaching the dimensions of natural ion channels:
K. Göpfrich, T. Zettl, A. E. C. Meijering, S. Hernández-Ainsa, S. Kocabey, T. Liedl & U. F. Keyser, DNA-tile structures lead to ionic currents through lipid membranes. Nano Letters, 2015. http://pubs.acs.org/doi/abs/10.1021/acs.nanolett.5b00189
2014
DNA-based membrane pores exhibit voltage-dependent conductance states, reminiscent of gating observed for natural membrane pores:
A. Seifert*, K. Göpfrich*, J. R. Burns, N. Fertig, U. F. Keyser & S. Howorka, Bilayer-spanning DNA nanopores with voltage-switching between open and closed state. ACS Nano, 2014 (*equal contribution). http://pubs.acs.org/doi/abs/10.1021/nn5039433
2013

Two porphyrin-tags anchor a simple DNA nanopore in the lipid membrane and serves as fluorescent dyes at the same time:
J. R. Burns, K. Göpfrich, J. W. Wood, V. V. Thacker, E. Stulz, U. F. Keyser & S. Howorka, Lipid-bilayer-spanning DNA nanopores with a bifunctional porphyrin anchor. Angewandte Chemie International Edition, 2013. http://onlinelibrary.wiley.com/doi/10.1002/anie.201305765/abstract
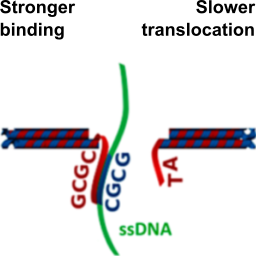
We modify solid-state nanopores with DNA origami to control their pore size and the positioning of binding sites for specific analytes:
S. Hernández-Ainsa, N. A. W. Bell, V. V. Thacker, K. Göpfrich, K. Misiunas, M. E. Fuentes-Perez, F. Moreno-Herrero & U. F. Keyser, DNA origami nanopores for controlling DNA translocation. ACS Nano, 2013. http://pubs.acs.org/doi/abs/10.1021/nn401759r
The frequency of DNA translocation through the protein nanopore alpha-hemolysin is significantly enhanced at pH 6 compared to pH 8:
K. Göpfrich, C. V. Kulkarni, O. J. Pambos & U. F. Keyser, Lipid nanobilayers to host biological nanopores for DNA translocations. Langmuir, 2013. http://pubs.acs.org/doi/abs/10.1021/la3041506
